- the rationale behind the allocation of people to each of the new Genetic Families
There are 4 haplogroups represented in the project (E, G, I, and R). A haplogroup is simply a group of people with a similar genetic signature. There are 20 Y-DNA haplogroups altogether, named after the letters A through S. Each of these haplogroups can in turn be subdivided into smaller and smaller subgroups (or “subclades”). And all of these groups can be placed on an evolutionary tree (the haplotree) that summarises the evolution of the human Y-DNA signature from its earliest origins in Africa to its arrival in Europe about 45,000 years ago, and Ireland about 13,000 years ago.
Genetic Families can be considered the smallest subgroups of this evolutionary tree - the final twigs on the branches of the haplotree.
But to start with, let’s take a look at each of these major haplogroups in turn, and the genetic families that have been identified within each one.
Haplogroup E and G
The members belonging to these haplogroups are singletons (i.e. no close matches within the project) and have been allocated to the “Ungrouped (non-R)” category.
Haplogroup I
Of the 7 members in Haplogroup I, four have been grouped into 2 distinct genetic families and the remaining 3 are ungrouped singeltons.
I1-Genetic Family 1 (I1-GF1)
I1-Genetic Family 1 (I1-GF2)
Haplogroup R1a
The 2 members who belong to Hg R1a now form a distinct genetic family, namely R1a-Genetic Family 1 (R1a-GF1 for short).
Next week we’ll take a look at the largest haplogroup within the project, Hapolgroup R1b, and the 5 genetic families that belong to it.
Genetic Families can be considered the smallest subgroups of this evolutionary tree - the final twigs on the branches of the haplotree.
But to start with, let’s take a look at each of these major haplogroups in turn, and the genetic families that have been identified within each one.
 |
| The 20 Haplogroups on the Y-DNA Haplotree (from FTDNA) |
Haplogroup E and G
The members belonging to these haplogroups are singletons (i.e. no close matches within the project) and have been allocated to the “Ungrouped (non-R)” category.
Haplogroup I
Of the 7 members in Haplogroup I, four have been grouped into 2 distinct genetic families and the remaining 3 are ungrouped singeltons.
I1-Genetic Family 1 (I1-GF1)
- The 2 members of I1-GF1 both bear the surname Farrell.
- They differ from each other by a GD of 0/37 and 1/67, which indicates a very close relationship between the two individuals, possibly as close as second or third cousins.
- The TiP24 score [1] is 100%. In fact, their TiP Report suggests that there is a 95% chance that the common ancestor was born sometime within the previous 8 generations (which equates to sometime after about 1700 assuming 30 years between generations and a date of birth of the participants of about 1940).
- Possible “rare” marker values: none obvious
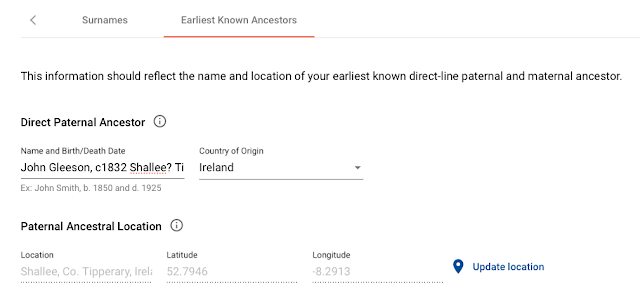
- The terminal SNP [2] for both these members is I-L205, placing them in the following subclade of the ISOGG Y-DNA tree 2015: I1a1b2 (see http://www.isogg.org/tree/ISOGG_HapgrpI.html)
- Their MDKA information does not list any specific locations but further research may reveal that their MDKAs were born in the same area.
- Given the estimated closeness of their relationship, these participants should share their genealogical data and try to ascertain who is their common ancestor or where he may have come from.
 |
| I1-GF1 and I1-GF2 (click to enlarge) |
I1-Genetic Family 1 (I1-GF2)
- The 2 members of I1-GF2 share the surname O’Farrell.
- They have been grouped together despite the fact that member 103146 appears to have some trouble with his results. Specifically, some marker values are missing (the multi-copy markers DYS459, DYS464, and CDYa & b). I’m assuming that this is a technical issue. If one ignores these marker values, the two haplotypes are identical and therefore have been grouped together.
- The TiP24 score for these two members is only 51% but this may be due to the technical error with the marker values (or alternatively they may be incorrectly grouped together). I will check with FTDNA and see if the error can be corrected. If the corrected data shows that the two haplotypes (i.e. genetic signatures) are identical, then these two individuals are probably very closely related and may share a common ancestor within the last 8 generations or so (i.e. since 1700).
- Possible “rare” marker values: none obvious
- The terminal SNPs for these two members are consistent, with P109 (subclade I1a1b1) being downstream of M170 (Hg I) - see http://www.isogg.org/tree/ISOGG_HapgrpI.html
- The country of origin of both these members MDKA (Most Distant Known Ancestor) is given as Ireland (see Results in Classic mode). However, all other MDKA data is missing completely for one member and there are no locations mentioned for the second member, so it is not currently possible to see if the MDKAs for these two members came from the same part of Ireland. Both members should update their MDKA data accordingly.
Haplogroup R1a
The 2 members who belong to Hg R1a now form a distinct genetic family, namely R1a-Genetic Family 1 (R1a-GF1 for short).
- Their surnames (Farr & Farrar) could well be variants of each other.
- They differ by a GD of 4/37.
- On the TiP calculator, the TiP24 score is 96.96% supporting the placement of these 2 members within the same genetic family.
- Possible “rare” marker values:
- DYS19 is 16 (occurs in only 10.6% of the general population, but 38% of the R1a population so this is not really rare)
- The terminal SNP’s for these two members are consistent, with Z93 (subclade R1a1a1b2) being downstream of M512 (subclade R1a1a) - see http://www.isogg.org/tree/ISOGG_HapgrpR.html
- The members of this group are probably not very closely related to each other as the TiP tool estimates a 90% chance of a common ancestor within the last 20 generations approximately (91.77% in fact). That would mean that there is a roughly 90% chance that the common ancestor was born some time after 1340 (if one allows 30 years per generation and a dob of the members of about 1940). Equally, there is a 10% chance that their common ancestor was born before 1340 (approximately). Nevertheless, these members should share their genealogical data and try to ascertain who is the common ancestor or where he may have come from. If one or both of them have an extensive pedigree then they might get lucky.
 |
| R1a-GF1 (click to enlarge) |
Next week we’ll take a look at the largest haplogroup within the project, Hapolgroup R1b, and the 5 genetic families that belong to it.
[1] The TiP24 score is the value
obtained from the TiP Report at 24 generations with the following settings: 1)
comparison set to the 37-marker level; 2) default settings (i.e. they do not
share a common ancestor more recently than 1 generations ago; display every 4 generations).
In this situation, the TiP Report is not being used to estimate the time to
most recent common ancestor (TMRCA) but rather as a more accurate estimate of
relative closeness than merely GD. This is because GD does not take into
account the variable mutation rates of markers whereas the TiP Report does.
This technique was developed by James Irvine and is used in his Clan Irwin
Surname DNA Study (https://www.familytreedna.com/public/irwin).
[2]
There are two types of marker on all chromosomes - SNP markers and STR markers. STR markers are
the row of numbers you see on the Results page. SNP markers are a different
type of marker and are used to subdivide members of a haplogroup into smaller
and smaller subgroups/subclades. The terminal SNP is the marker that identifies
the current end of a particular branch of the haplotree. More SNP markers will
be discovered in time that will identify additional (smaller) subgroups further
“downstream” from the current “terminal SNP”. In other words, the “terminal
SNP” changes over time as more markers are discovered and their position on the
haplotree is clarified.